Working with Molecules
This section describes GaussView's molecule building and viewing features.
These options enable you not only to construct new structures either atom-by-atom
or from fragments, but also to manipulate and examine previously-computed molecules.
Placing Elements and Fragments
The GaussView molecule building tools always operate on the active view
window. In general, GaussView begins and remains in its insert/replace
mode, unless one of the options described below explicitly calls for it to act
in a different manner. Clicking in an open area of the view window adds the
current atom or fragment to the window as a distinct fragment. Clicking on an
existing atom adds the current item to the existing structure at the indicated
point, usually replacing it (other options are discussed later).
The desired atom or fragment is chosen using one of the Element button, Groups
button, Rings button and Bio button and then selecting one of the choices from
the resulting palette. The current item always appears in the Current Fragment
area of the GaussView control window or the Active Fragment area of the
standalone Builder palette (see Figure 4). The standalone Builder palette is
opened with the View=>Builder menu item.
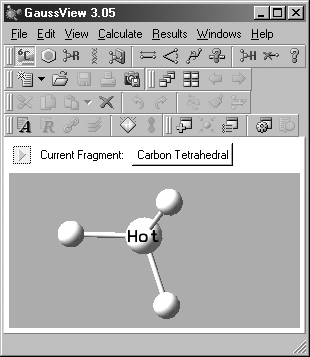
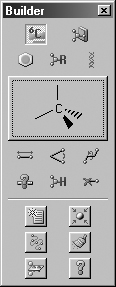
Figure 4. Builder Current/Active Fragment Displays
The selected atom or fragment appears in both the control window and on the
old-style Builder palette if it is visible (highlighted above). On the latter,
clicking on Active Fragment window opens the palette corresponding to the selected
fragment type icon above it. On the control panel and the detached Builder toolbar,
clicking again on a selected fragment type icon opens its palette, as does clicking
on the Current Fragment display field (labeled "Carbon Tetrahedral" here).
The Element Palette
The Select Element window allows you to select an element from the periodic
table. The various possible coordination patterns for that element are shown
at the bottom of the window (see Figure 5). The default element is tetrahedral
carbon.
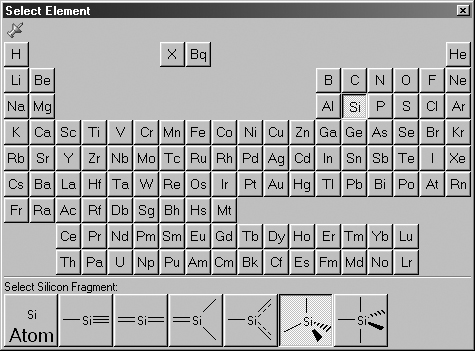
Figure 5. The Select Element Palette
The available connectivity variations appear along the bottom of the palette.
The Group Palette
The Select R-Group Fragment window allows you to select from a series
of AM1-optimized functional group fragments. It is illustrated in Figure 6.
The default group is carbonyl (formaldehyde).
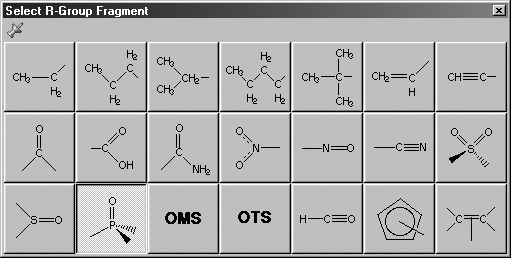
Figure 6. Select R-Group Fragment Palette
Use this palette to select the desired functional group.
The Rings Palette
The Select Ring window allows you to select from a series of AM1-optimized
ring structures. It is illustrated in Figure 7. The default ring is benzene.
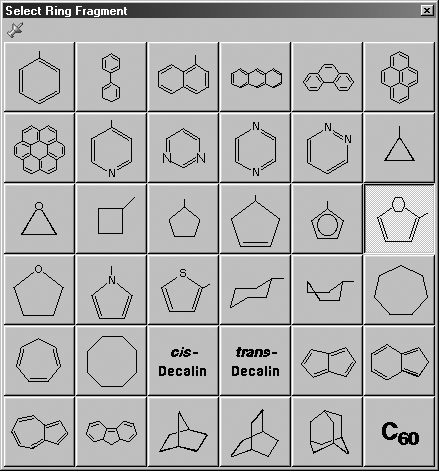
Figure 7. The Select Ring Fragment Window
Use this palette to select the desired ring system.
Note that several of the fragments use non-traditional bonding. These fragments
include a five-member ring with an open bond perpendicular to the plane of the
ring. This is a cyclopentadienyl ligand for use in constructing organometallic
systems.
The Bio Palette
The Biological Fragments dialog presents a set of AM1-preoptimized fragments
useful to biological researchers: the amino acids and various DNA bases (see
Figure 8). The default Bio fragment is Alanine Central Residue.
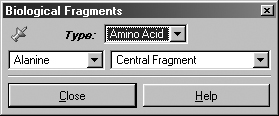
Figure 8. Selecting Biological Fragments
Use the various fields in this window to select amino acids and DNA bases.
The Type field allows you to select either Amino Acid or Nucleoside.
When Amino Acid is selected, the left popup menu below the Type
field can be used to select the desired amino acid, and the right popup menu
in that line allows you to specify the structural form as Central Fragment,
Amino-Terminal Fragment or Carboxyl-Terminal Fragment (the last
two are the N-terminated and C-terminated forms, respectively).
When the Type field is set to Nucleoside, the left popup menu
below the Type field can be used to select the desired DNA base, and
the right popup menu in that line allows you to specify the structural form
as Central Fragment, C3'-Terminal Fragment, C5'-Terminal Fragment
or Free Nucleoside.
Making Palettes Sticky
Unlike in earlier versions of GaussView, the fragment palettes currently
in GaussView can be used in a modal (dialog box behavior) or amodal (floating
palette behavior) manner. The thumbtack icon in the upper left corner of the
window shows whether the palette will close when another one is selected (or
a non-building context is entered). The pushed-in tack indicates that the palette
is "tacked" to the desktop (i.e., "sticky"): . The upward pointing tack similarly
indicates modal operation: .
Undo and Redo
The Edit=>Undo and Edit=>Redo menu paths may be used to reverse or reinstate
a previous molecule modification or other action. The Undo button and the Redo
button may be used for the same purposes.
Previous Next





