The Atom List Editor
The Atom List Editor window is displayed when you select the Edit=>Atom
List menu item or click on the Atom List button. This tool presents the coordinates
and other parameters for the molecule in the active view window in an editable
spreadsheet format. The window's default configuration is illustrated in Figure
28.
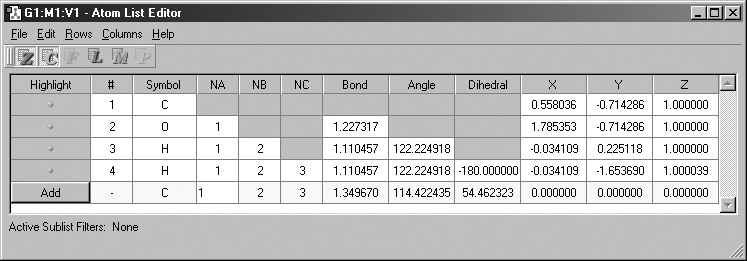
Figure 28. The Atom List Editor
Any of the molecular parameters can be edited in this spreadsheet-like windows.
Atoms may be selected by clicking on the dot in the leftmost column (and deselected
by clicking it a second time). The corresponding atoms will also be selected/deselected
in the molecule's view window. You can add additional atoms by scrolling to
the bottom of the windows, and then clicking the Add button. This inserts an
new row into the list (filled with default values, as above).
The default configuration of this window displays the molecule's Z-matrix and
Cartesian coordinates. Different data can be selected for display using the
toolbar buttons. From the left, they control Z-matrix coordinates, Cartesian
coordinates, fractional coordinates, ONIOM layer assignments and related settings,
Molecular Mechanics atoms types and other data, and PDB file data about the
residue and chain. Columns corresponding to the final button are read-only.
Figure 29 illustrates the additional columns that may be present in this window.
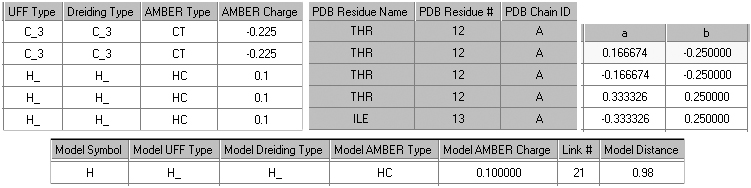
Figure 29. Additional Atom List Editor Columns
Beginning at the upper left and moving clockwise, these columns are controlled
by the MM Data button, the PDB Info button, the Fractional Coords button, and
the ONIOM Layers button.
The various menu items in this window have the following functions
-
File=>Print: Print the atom list table.
-
File=>Exit: Exit from the Atom List editor.
-
Edit=>Sublist Filters: Define, modify and
remove filters which determine which atoms are displayed and also may be
used to delete a group of atoms in a single operation. Filters may be based
on element type, ONIOM layer, and atom number ranges and/or lists. Multiple
filters may be in use at the same time.
-
Edit=>Delete Atoms: Delete atoms. The choice of which atoms to delete
is based on the selection made from the submenu: All (all atoms), Selected
(only selected atoms), Hydrogen (all hydrogen atoms), Redundant (removes
duplicate rows), Sublist (atoms selected by the active sublist filter),
Non-Sublist (atoms not selected by the sublist filter).
-
Edit=>Sort Atoms: Rearrange rows according to the choice made on the
submenu: Hydrogens Last (place all hydrogen atoms at the bottom of the list),
Reorder by Connectivity (standard Z-matrix order).
-
Edit=>Z-Matrix=>Reconnect: Regenerate the Z-matrix.
-
Edit=>Optimization Flags: Not applicable to Gaussian.
-
Edit=>PBC=>Trim Contents: Atoms that are removed are determined by
the selection from the submenu: Delete All Atoms Outside Cell (remove atoms
not inside the unit cell), Translate All Atoms Inside Cell (all atoms outside
the cell boundary are translated to equivalent positions inside the cell
and then redundant atoms are deleted).
-
Edit=>Add Hydrogens: Add hydrogen atoms to the structure (typically
used with PDB files).
-
Edit=>Update MM Types: Regenerate Molecular Mechanic atoms types for
the molecule.
-
Rows=>Sort By: Sort atom list rows, The criterion is selected on the
submenu: Tag (sort by atom number), or Symbol (sort by element type).
-
Rows=>Sublist: Display atoms in or not
in the active sublist (depending on the submenu item that is chosen).
-
Help: Obtain help for this window.
The Columns menu allows you to specify which columns appear in the table in
more detail
-
Columns=>Z-Matrix: Toggle display of the Z-matrix coordinate or connectivity
data, depending on the submenu item chosen.
-
Columns=>Cartesian Coordinates: Toggle display of Cartesian coordinates.
-
Columns=>Optimization Flags: Not applicable to Gaussian.
-
Columns=>Fractional Coordinates: Toggle display of fractional coordinate
a, b, or c, depending on the submenu item chosen.
-
Columns=>ONIOM Data: Toggle display of various ONIOM-related columns,
selected from the submenu: Layer (layer assignment), Freeze (optimization
freeze parameter), Model Symbol (element to use as a link atom), Model MM
Types (submenu allowing you to select which link atom MM type(s) are displayed),
Link # (atom to which the link atom is bonded in the model system), Model
Distance (link atom bond distance in the model system). The Model MM Types
submenu allows you to toggle the display of the link atom MM type for UFF,
Dreiding, and/or AMBER, as well as the AMBER Charge. Note that the display
of MM types for the atom itself are controlled by the next item.
-
Columns=>MM Types: Toggle the display of the MM types for the atoms
in the molecule. The UFF, Dreiding, and AMBER types can be individually
selected, as can the AMBER Charge. The AMBER Fragment does not apply to
Gaussian.
-
Columns=>PDB Data: Select PDB items to display, as selected from the
submenu of Residue Name, Residue #, and Chain ID.
The Atom List Editor's Sublist Filters
The Sublist Filter window is displayed when you select the Atom List
Editor's Edit=>Sublist Filters menu item. This tool allows you
to specify complex filters which specify which atoms appear or do not appear
in the Atom List Editor's display. It is illustrated in Figure 29a.
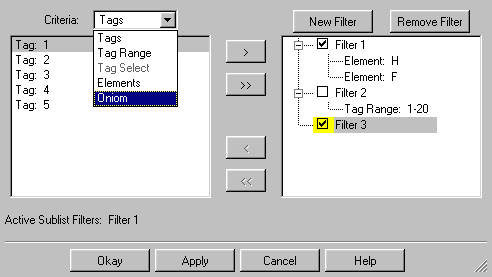
Figure 29a. Specifying Sublist Filters
The left list box is used to select criteria when editing filters, and the
right list box displays currently defined ones. Active filters are checked,
and the highlighted line is the criterion that is being defined or edited. Here,
we are just starting to add a third filter to the set. All active filters only
effect the Atom List Editor display when the Apply button is pressed. The particular
effect that results depends on the items that are checked in the main window's
Rows=>Sublist submenu.
The various items in this dialog will now be considered individually:
- The Criteria menu is used to set up filter conditions, selecting
atoms on various grounds. The currently available conditions for the specified
criterion type appear in the left hand list box.
-
The New Filter button adds a new filter to the right hand list box,
and the Remove Filter item removes the entire currently-selected
filter.
-
Individual items are moved between the two lists using the >
and < buttons. The former adds the current criterion to the selected
filter, and the latter removes the selected criterion in the filter list
from its filter.
-
The >> and << buttons move all criteria in the
corresponding list.
-
The Apply button applies the currently selected filters. Note that
all filter conditions are ANDed.
- The Okay and Cancel buttons both close the dialog, retaining
and discarding filter changes respectively.
Filter Effects
The Atom List Editor's Rows=>Sublist submenu contains two items:
Show Sublist Atoms and Show Non-Sublist Atoms. Each item can be
individually checked or unchecked, indicating whether the corresponding rows
are shown when a sublist filter is active. If no filter is active, then this
submenu is disabled.
Previous Next


