Figure 26. GaussView Fields in the Save File Dialog
GaussView adds several fields to the standard system Save dialog. Molecules can be written in several formats: as a Gaussian input file, in MDL Mol file format, and in Sybyl Mol2 file format.
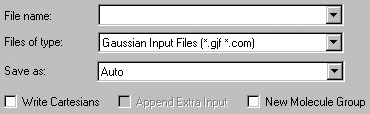
The File=>Save menu item and the Save button allow you to save the current model to an external file or set of files. These items use the system's Save dialog, to which GaussView adds some additional fields (see Figure 26).

Figure 26. GaussView Fields in the Save File Dialog
GaussView adds several fields to the standard system Save dialog. Molecules
can be written in several formats: as a Gaussian input file, in MDL Mol file
format, and in Sybyl Mol2 file format.
These fields have the following meanings
Files of type: Acts as a filter, selecting which files are displayed in the dialog's file list.
Save as: Forces the saved file to be written as the selected type, regardless of the file extension that it is assigned. The default is Auto, which selects the output file format based on its extension.
Write Cartesian Coordinates: If checked, the molecular structure is written in Cartesian coordinates in the saved file. By default, a Z-matrix is written.
Append Extra Input Data: This control is active for models which were read in from Gaussian input files. If there was additional input present in the file, checking this box will cause it also to be included in the output file.
Create New Molecule Group: If checked, GaussView also creates a new molecule group containing the saved molecule.
Within a molecule group, the current model is saved. There is no way to save all of the models within a molecule group to a single file or to a group of files in the current program version.
Previous Next